Structure, Mechanism and RNA Therapeutics
The advent of RNA therapeutics
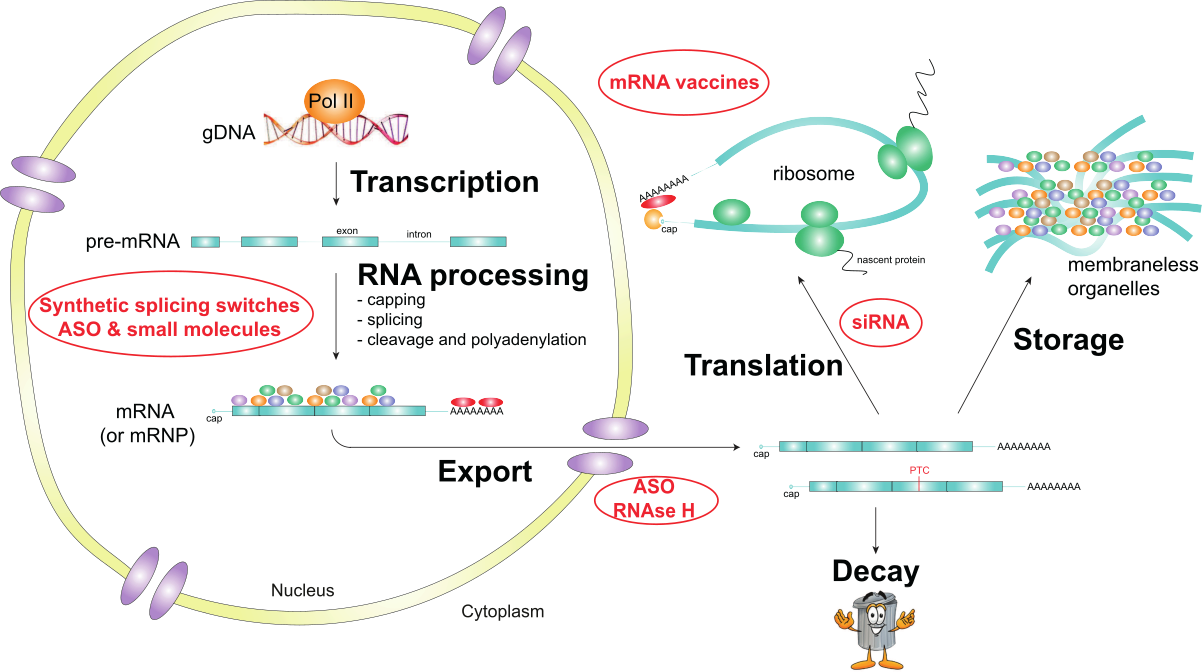
In eukaryotes, the nuclear envelope breaks the physical connection between transcription and translation observed in prokaryotes. The nuclear envelope also creates a time and a space for RNA processing, impacting mRNA localization, translation, and decay. During transcription, pre-mRNA undergoes capping, splicing, cleavage, and polyadenylation in the nucleus before being exported to the cytoplasm where it will be translated, stored or degraded. The regulation of RNA processing shapes genetic information and, therefore, gene expression. Altered RNA metabolism has been linked to human inherited diseases and cancers. RNA therapeutics, including mRNA vaccines, siRNA, antisense oligonucleotides, and small molecules, aim to restore physiological RNA processing and gene expression patterns. Our research group aims to investigate the fundamental basis of RNA therapeutics acting on nuclear RNA processing and develop this field of medicinal chemistry.
Fundamental basis of pre-mRNA splicing regulation
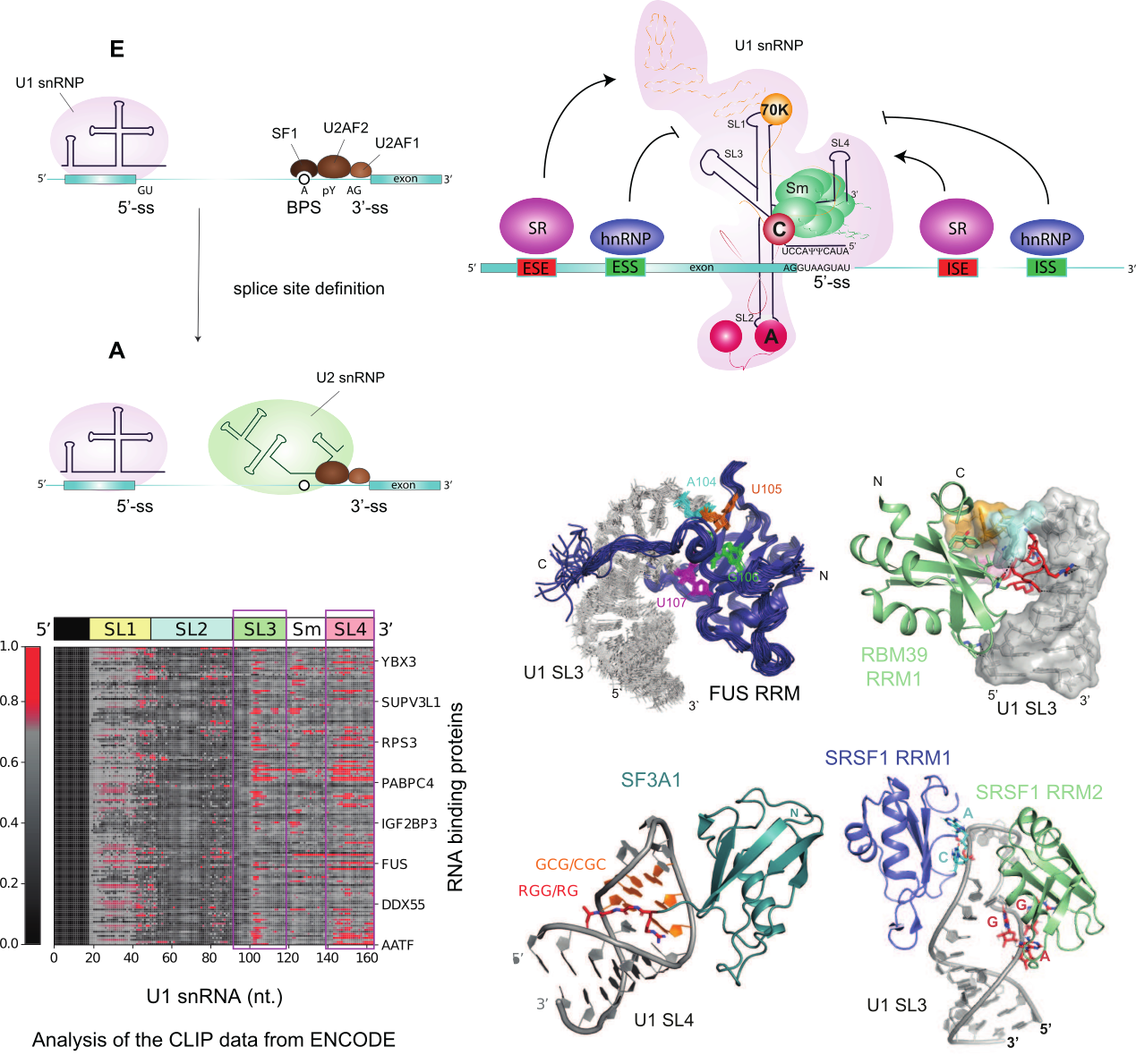
RNA splicing is a crucial process that involves the removal of introns and the linking of exons to form messenger RNA, thus playing a major role in shaping the transcriptional output. Alternative splicing (AS) allows eukaryotic cells to generate multiple mRNAs from a single precursor mRNA. The recognition of the 5’-splice site by the first particle of the spliceosome U1 snRNP is a major hotspot for RNA splicing regulation. 5’-splice site definition is influenced by the 5’-splice site sequence complementarity with the U1 snRNA 5’-end, its accessibility, and the presence of cis-regulatory RNA elements nearby. Our research group is investigating how splicing factors communicate with U1 snRNP to help recognize weak 5’-splice sites. We use an in vitro reconstituted U1 snRNP (Campagne S. et al., Nucleic Acids Research 2021) to probe the interactions between splicing factors and U1 snRNP. Our recent studies show that the solvent-accessible stem loops, SL3 and SL4, play a significant role in mediating these interactions (Campagne S. et al., Nat. Communications 2020, PNAS 2022). Using CLIP data released by the ENCODE project, we identified over 80 splicing factors capable of interacting with these stem loops and regulating gene expression (Campagne S. et al., EMBO J 2022). Our aim is to determine the structural basis of these interactions and understand the functional consequences of trans-splicing factor competition on these sites.
RNA metabolism and blood cell fitness
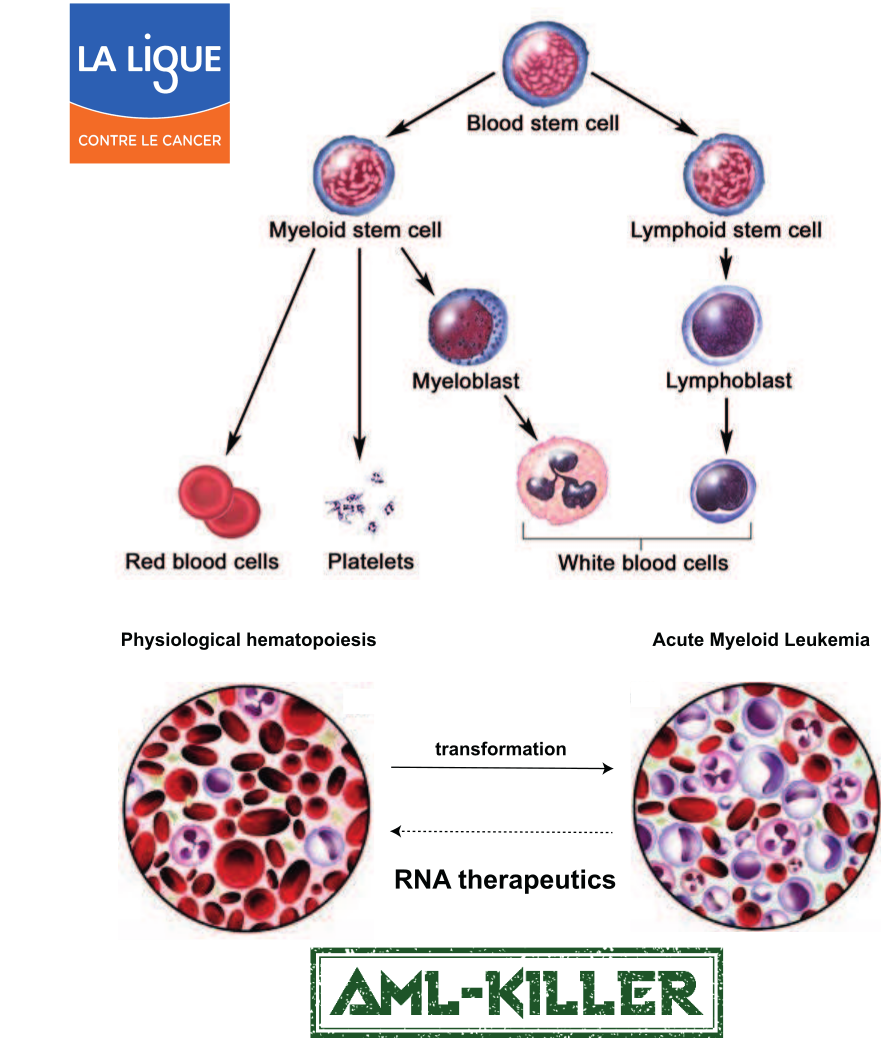
Blood cell progenitors reside in the bone marrow, where the first asymmetric division gives rise to myeloid and lymphoid lineages. Following a long process of differentiation involving cell death of mis-differentiated progenitors, B and T cells emerge from the lymphoid lineage. Our research focuses on understanding the molecular mechanisms that trigger the Ca2+-activation of an endonuclease that promotes the apoptosis of mis-differentiated cells in the lymphoid lineage, as loss of function of this endonuclease has been linked with various cancers. In addition, we are developing RNA therapeutics to combat Acute Myeloid Leukemia (AML), a rare yet deadly cancer associated with a 5-year survival rate of less than 30%. Our AML-killer project was funded by the federal council of La Ligue contre le cancer (through the call “RNA therapeutics”) and aims to target an RNA binding protein essential for AML cell survival, independent of the adaptor protein used by current approaches.
Fundamental basis for small molecule splicing modifiers
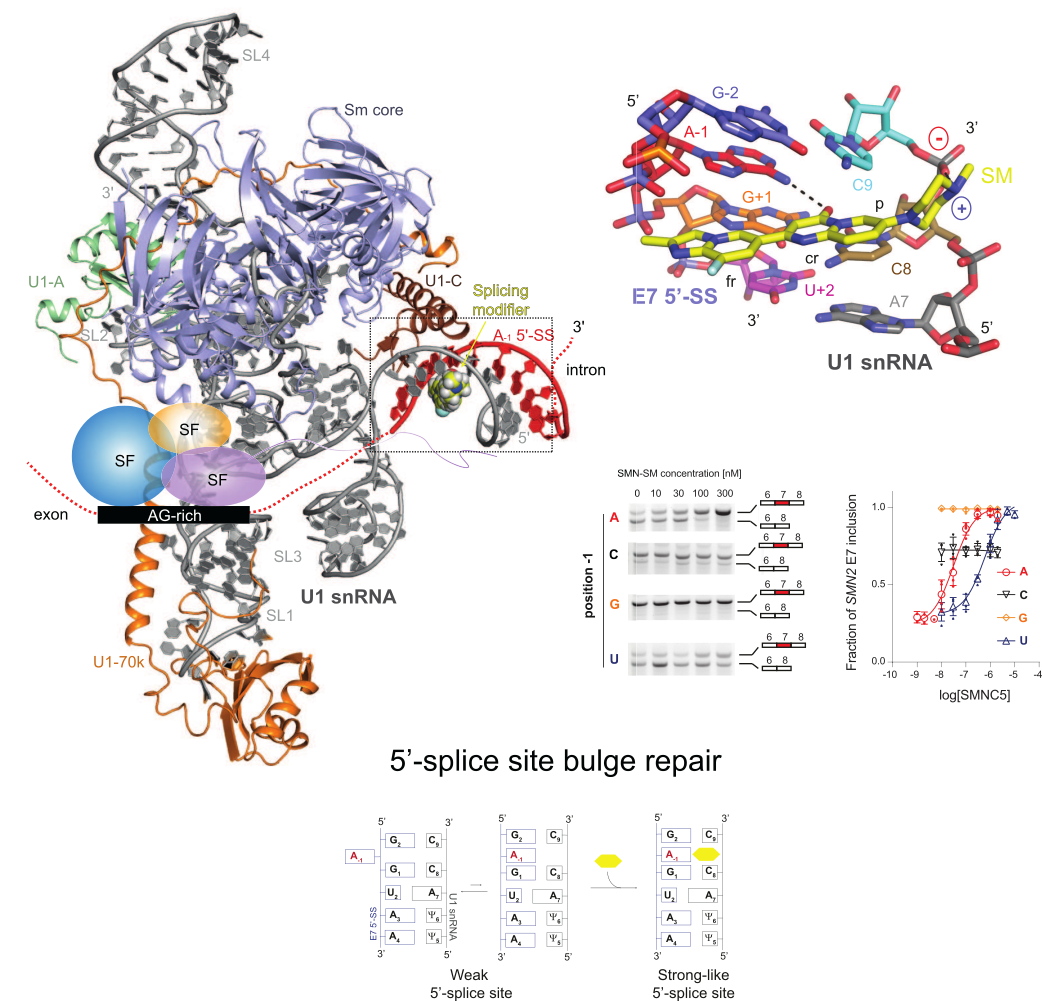
RNA splicing correction is an emerging area for drug discovery, having already yielded significant therapeutic breakthroughs. During my previous work, I have discovered the mechanism of action of the first small molecule that specifically corrects SMN2 splicing. The SMN2 splicing modifiers facilitate spliceosome assembly on a limited set of exons by acting as molecular glue at the spliceosome/RNA interface, repairing a bulge at the position -1 of the 5’-splice site. Although we have identified their mechanism of action, gene selectivity of the small molecule splicing modifiers is still not fully understood. Our lab combines structural and functional approaches to understand the fundamental mechanisms of small molecule splicing modifiers in order to develop new splicing modifiers with different gene specificities. We strongly believe that small molecule splicing modifiers could provide innovative therapeutic solutions for human diseases caused by mutations in the 5’-splice site.